The following code can be used to calculated a confidence interval using simulation techniques. This is in the context of class for October 2nd.
Packages
Warning: package 'ggplot2' was built under R version 4.3.3
Warning: package 'tidyr' was built under R version 4.3.3
Warning: package 'readr' was built under R version 4.3.3
Warning: package 'dplyr' was built under R version 4.3.3
Warning: package 'stringr' was built under R version 4.3.3
Warning: package 'lubridate' was built under R version 4.3.3
── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.5.1 ✔ tibble 3.2.1
✔ lubridate 1.9.3 ✔ tidyr 1.3.1
✔ purrr 1.0.2.9000
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
Warning: package 'tidymodels' was built under R version 4.3.3
── Attaching packages ────────────────────────────────────── tidymodels 1.2.0 ──
✔ broom 1.0.5 ✔ rsample 1.2.1
✔ dials 1.2.1 ✔ tune 1.2.0
✔ infer 1.0.7 ✔ workflows 1.1.4
✔ modeldata 1.3.0 ✔ workflowsets 1.1.0
✔ parsnip 1.2.1 ✔ yardstick 1.3.1
✔ recipes 1.0.10
Warning: package 'dials' was built under R version 4.3.3
Warning: package 'scales' was built under R version 4.3.3
Warning: package 'infer' was built under R version 4.3.3
Warning: package 'modeldata' was built under R version 4.3.3
Warning: package 'parsnip' was built under R version 4.3.3
Warning: package 'recipes' was built under R version 4.3.3
Warning: package 'rsample' was built under R version 4.3.3
Warning: package 'tune' was built under R version 4.3.3
Warning: package 'workflows' was built under R version 4.3.3
Warning: package 'workflowsets' was built under R version 4.3.3
Warning: package 'yardstick' was built under R version 4.3.3
── Conflicts ───────────────────────────────────────── tidymodels_conflicts() ──
✖ scales::discard() masks purrr::discard()
✖ dplyr::filter() masks stats::filter()
✖ recipes::fixed() masks stringr::fixed()
✖ dplyr::lag() masks stats::lag()
✖ yardstick::spec() masks readr::spec()
✖ recipes::step() masks stats::step()
• Use tidymodels_prefer() to resolve common conflicts.
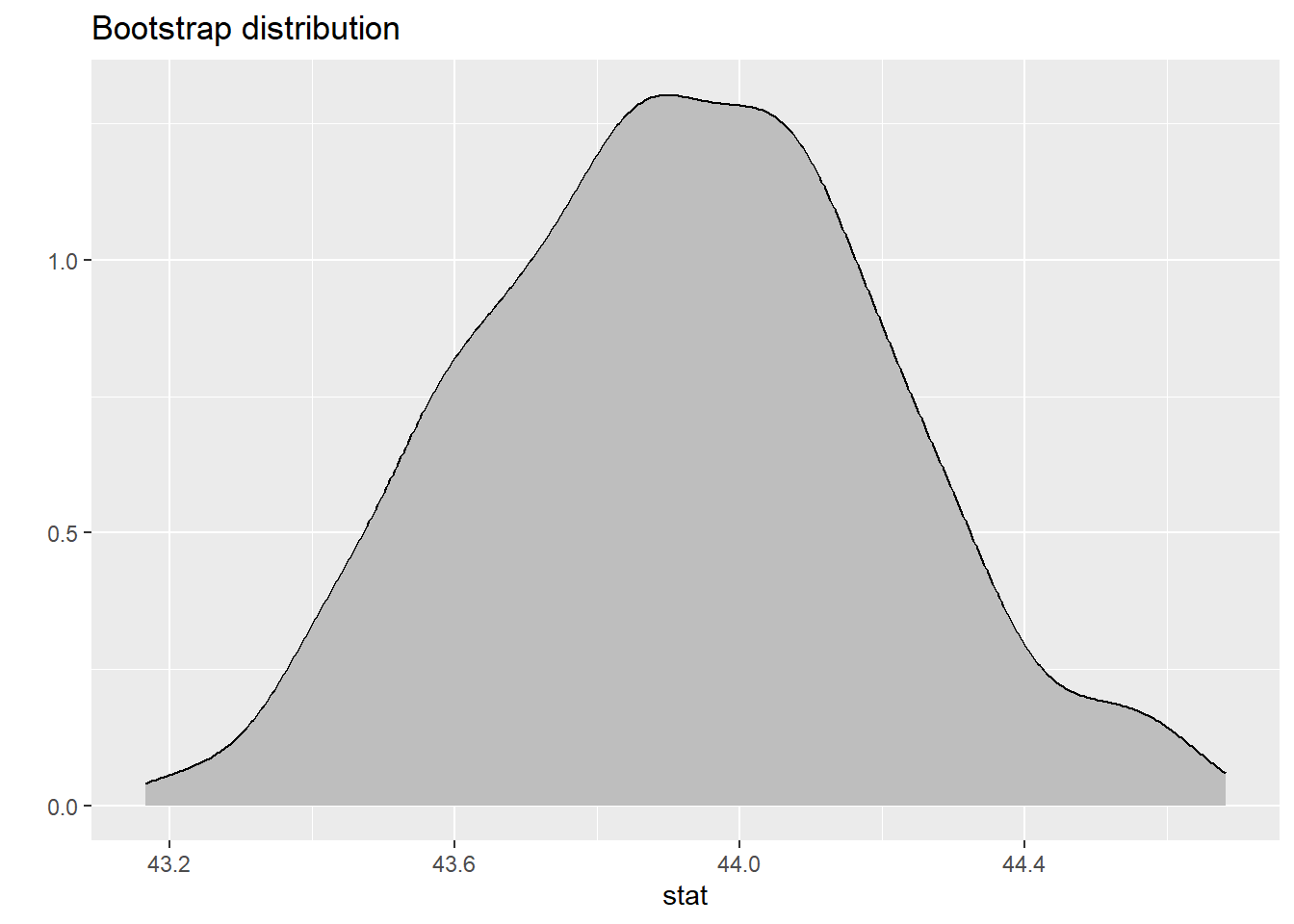
Because we can assume that the assumptions are valid for the central limit theorem, we are going to use simulation based techniques to estimate the true mean bill length of penguins.
See the slides for the steps on how the simulated sampling distribution is created. Below is the code to create this distribution.
The Process
set.seed ( 12345 ) boot_df <- penguins |> specify ( response = bill_length_mm ) |>
generate ( reps = 1000 , type = "bootstrap" ) |>
calculate ( stat = "mean" )
Warning: Removed 2 rows containing missing values.
The Calculation
# A tibble: 1 × 2
lower upper
<dbl> <dbl>
1 43.4 44.5